 Portrait of a man in red chalk (1512). “/>
Portrait of a man in red chalk (1512). “/>Guadalupe Piñar et al.
Microbiomes are in vogue, even in the preservation of art, in which the study of microbial species that gather in works of art can lead to new ways to slow down the deterioration of invaluable works of art, as well as the potential to dismantle forgeries. For example, scientists analyzed microbes found on seven of Leonardo da Vinci’s drawings, according to a recent paper published in the journal Frontiers in Microbiology. And in March, scientists at the J. Craig Venter Institute (JCVI) collected and analyzed swabs taken from centuries-old art in a private collection housed in Florence, Italy, and published their findings in the journal Microbial Ecology.
The researchers behind the previous paper in March were JCVI geneticists who collaborated on the Leonardo da Vinci DNA project in France. The paper was based on a previous study looking for microbial signatures and possible geographic patterns in hair collected from people in the District of Columbia and San Diego, California. They concluded from that analysis that microbes could be a useful geographical signature.
For the March study, JCVI geneticists took microbial swabs from Renaissance-style pieces and confirmed the presence of so-called “positive oxidase” microbes on painted surfaces of wood and canvas. These microbes eat the compounds found in paint, glue and cellulose (found in paper, cloth and wood), which in turn produce water or hydrogen peroxide as by-products.
“It is possible that such by-products influence the presence of mold and the overall rate of deterioration,” the authors noted in their paper. Although previous studies have attempted to characterize the microbial composition associated with the decomposition of works of art, our results summarize the first large-scale genomics-based study to understand the microbial communities associated with aged works of art.
As an added bonus, they found that they could discern between microbial populations on different types of materials. Specifically, stone and marble art has encouraged more diverse populations than paintings, possibly due to “the porous nature of stone and marble that houses additional organisms and potentially moisture and nutrients, along with the likelihood of biofilm formation,” they wrote. In contrast, oil paintings provided weaker nutrients for microbes to metabolize.
The authors acknowledged the small size of the sample, but nevertheless concluded that microbial signatures could be used to differentiate works of art according to the materials used. As always, more research is needed. “Of particular interest would be the presence and activity of enzymes that degrade oil,” the authors wrote. “Such approaches will lead to a full understanding of the organism (s) that are responsible for the rapid decay of works of art while potentially using this information to target these organisms to prevent degradation.”
Swabing renaissance art
-
Collage of various works of art taken for a March work by geneticists at the J. Craig Venter Institute. Circles indicate buffer zones on each art sample
JCVI
-
Manolito G. Torralba and colleagues used small, dry polyester swabs to easily collect centuries-old, Renaissance-style microbes in a private collection house in Florence, Italy.
Jesse Ausubel
Guadalupe Piñar and her team at the University of Natural Resources and Life Sciences in Vienna, Austria, collaborated with curators at the Central Institute for Pathology of Archives and Books (ICPAL) to analyze the microbiome of Leonardo da Vinci’s drawings. Last year, Piñar et al. relied on microbiome analysis to study the storage conditions of three statues taken from smugglers, as well as to identify their possible geographical origins. Earlier this year, they analyzed the 1,000-year-old parchment microbiome, from which they could deduce the animals whose skins were used to make the parchments.
For this latest work, Piñar’s team used a third-generation sequencing method known as Nanopores, which uses protein nanopores embedded in a polymeric membrane for sequencing. It comes with a portable, pocket device, MinION, which makes it ideal for cultural heritage studies. For the drawings of Leonardo, Piñar et al. combined Nanopore sequencing with a genome-wide amplification protocol.
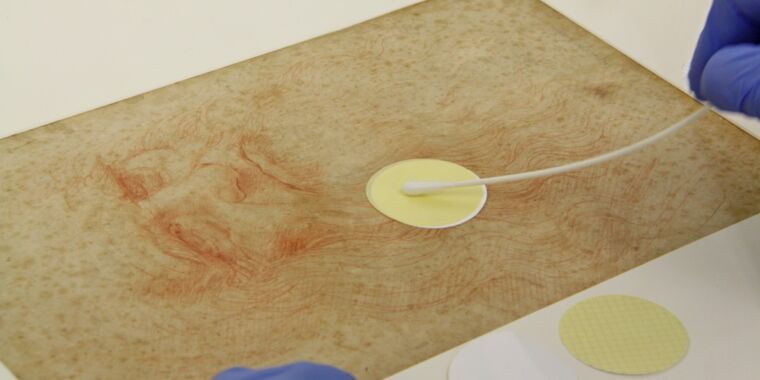
ICPAL preservatives have used a delicate, non-invasive sampling method (e.g., suction filtration) to collect dust particles, microbial cells, and other debris from small surfaces on both sides. Right and by of each drawing. The DNA was then extracted, amplified and sequenced. The Austrian team used optical microscopy to imagine features of interest in all seven drawings and scanning electron microscopy (SEM) to analyze all the micro-objects gathered from the drawings.
A “biological pedigree”
-
Leonardo da Vinci’s drawings were analyzed in a study by Austrian and Italian scientists.
Guadalupe Piñar et al.
-
Sampling of the microbiome from Leonardo da Vinci Study of curtains for a figure on the knee (ca. 1475)
Guadalupe Piñar et al.
-
Of Leonardo da Vinci Bitte’s man.
Guadalupe Piñar et al.
-
Insect droppings appear as waxy brown inlays on the fibers of the L4 pattern (Studies of the front legs of a horse).
Guadalupe Piñar et al.
-
SEM images of the surface of a membrane used for sampling the surface of drawing L3 (Nudes for the battle of Anghiari).
Guadalupe Piñar et al.
Each drawing had its own unique microbiome – an “independent molecular profile or biological pedigree”. Dar Piñar et al. were surprised to find that, in general, bacteria dominated the fungi in the microbiomes of the drawings, contradicting the widespread belief that fungi would be more dominant, given their greater potential for colonization on paper. The researchers did not detect any visible damage to the drawing other than the fox spots (small yellow-brown spots or spots).
Many of these bacteria are commonly found in human microbiomes, suggesting that they found their way into drawings while they were being manipulated during restoration – although one might speculate whether it came from the artist himself. (The authors note that dust bacteria could “stay in suspension” for long periods of time.) Other bacteria were typical of insect microbiomes and could have been introduced long ago by flies that deposited excrement on drawings. These excrements appeared after imaging analysis as waxy brown inlays in the fibers.
The Austrian / Italian team failed to definitively conclude whether any of the microbial contaminants date back to Leonardo’s time. It seems more likely that human microbial elements are due to more recent restoration work. This is most likely the case for the designation designated L4 (Studies of the front legs of a horse) in particular, which showed the lowest biodiversity and the heaviest contamination from human DNA. The authors make an additional hypothesis that the recipe Leonardo used – “a preparative layer made with powdered chicken bones in the form of powder, white lead, indigo … mixed with animal gelatin” – could have interfered with the preservation of the microbiome L4, so only recent DNA remained.
But Piñar insists that the possibility of tracking this type of data is still extremely valuable. “The sensitivity of the Nanopore sequencing method provides an excellent tool for monitoring art objects. It allows the evaluation of microbiomes and the visualization of their variations due to harmful situations,” she said. “It can be used as a bio-archive of the history of objects, providing a kind of fingerprint for current and future comparisons.”
DOI: Microbian Ecology, 2020. 10.1073 / pnas.1802831115
DOI: Frontiers in Microbiology, 2020. 10.1007 / s00248-020-01504-x (About DOI).